Chronic lymphocytic leukemia (CLL) is the most common clonal malignancy found in the elderly.1 This disease leads to uncontrolled proliferation of CD5+/CD19+/CD23+ lymphocytes, which results assembly of small and mature lymphocytes in the blood, bone marrow, lymph nodes, and spleen.2 The characteristics of the disease include an abnormal increase in white blood cell counts, infiltration of leukemic cells in the bone marrow, lymphadenopathy, and splenomegaly.3,4 The clinical proceeding of patients with CLL is considerably heterogeneous ranging from indolent to aggressive state.5 The pathogenesis of CLL is complex and a variety of environmental, genetic, and epigenetic alterations have been described.6 Micro-RNAs are small non-coding RNA and act as a regulator of gene transcription. Altered expression of micro-RNAs is a hallmark of various cancers, including CLL.7,8 Various studies have shown that micro-RNAs are involved in the pathogenesis, progression, and prognosis of different tumors.9 Some micro-RNAs acts as tumor suppressors and silencing by epigenetic mechanisms such as DNA methylation or histone de-acetylation may contribute to the development of CLL.10 MiR-129-2 is a negative regulator of the sex-determining region Y-box 4 (SOX4), an SRY-related oncogene, which is involved in the regulation of embryonic development and the determination of cell fate. SOX4 is overexpressed in several types of cancer and may play an important role in the metastasis and tumor progression.11 Silencing of miR-129-2 by DNA methylation has been reported in various tumors.12–14 However, conflicting results have been reported regarding the hematopoietic malignancies.15 We investigated the DNA methylation status of miR-129-2 in a group of CLL patients compared with a well-defined control group. We also investigated the correlation of miR-129-2 with some clinical and laboratory characteristics of patients with CLL.
Methods
The studied population encompassed 100 individuals: 50 CLL patients referred to the hematology-oncology research center of Shariati Hospital in Tehran, Iran, and 50 healthy matched controls. Patients were aged over 50 years, and their disease was confirmed by morphologic features and flow cytometry analysis (CD5+, CD19+, and CD23+).
Patients with CLL who also had an infectious disease or other medical comorbidities (hepatitis C and B, HIV, cardiovascular disease, myocardial infarction, and diabetes), or who had been treated for cancer in the last five years were excluded from the study. The control subjects were selected from individuals referred to for checkup examination to the Shariati Hospital, Tehran, Iran. The exclusion criteria for control subjects were the presence of any acute and chronic disease, febrile and inflammatory or underlying disease, and taking specific medicines or immunosuppressive drugs. All participants signed an informed consent form. The study was approved by the ethical committee of the High Institute for Research and Education in Transfusion Medicine (Ethical code: IR. TMI. REC. 1396. 024), Tehran, Iran.
About 5–10 mL of blood was collected in EDTA anti coagulated falcon tubes from the patients. After the completion of complete blood counts and flow cytometry tests, the rest of the samples were stored at -20 ºC until molecular analysis.
DNA was isolated from peripheral blood using a standard salting-out method.16 Concentration and purity of extracted DNA were measured in a Nanodrop spectrophotometer (NanoDrop® ND-1000, Thermo Fisher Scientific). The EpiTect Fast Bisulfite Kit (Qiagen, Germany) was used to treat approximately 2 µg of extracted DNA by sodium bisulfite during, which the unmethylated cytosines converted into the corresponding uracils, while the methylated cytosines remained unchanged in their positions. Sodium bisulfite modified DNA was stored at -20 ºC until use.
We used methylation-specific PCR (MSP) to assess the methylation status of the miR-129-2 gene in CLL and control patients. Primer sequences for the methylation analysis of miR-129-2 gene are given below.
Unmethylated reaction: F:5’ TTT AGT TTG TAT TAA TGA GTT GG TG 3’, R: 5’ AAA TCT CTA AAC AAA TAC AAT TC AA 3’.
Methylated reaction: F: 5’ TTT TAG TTC GTA TTA ATG AGT TG GC 3’, R: 5’ GAA TCT CTA AAC AAA TAC AAT TC GA 3’.
Touchdown MSP analysis was done in a Flex cycler2 thermal cycler (twin 96 G 5.4, Analytik Jena, Germany) using a hot start master mix (Ampliqon, Denmark), as previously described with slight modification.17 The cycling condition included 31 cycles with an initiation annealing temperature of 70 ºC for 30 seconds decreased by 0.5 ºC for each cycle followed by additional 21 cycles with an annealing phase of 55 ºC for 30 seconds. Following the completion of the MSP reaction, the electrophoresis of amplified products on a 2.5% agarose gel revealed a 169 and 168 bp for the methylated and unmethylated state, respectively.
The numerical values presented as mean±standard deviation were compared using the Student’s t-test or ANOVA test. The chi-square test or Fisher’s exact tests was performed to assess the differences between categorical variables. We used binary logistic regression analysis to evaluate the independent effect of each risk factor on the methylation status of miR-129-2 in CLL patients. Statistical analysis was performed using SPSS Statistics (SPSS Inc. Released 2009. PASW Statistics for Windows, Version 18.0. Chicago: SPSS Inc.) with a statistical significance level of p < 0.050.
Results
The mean age of CLL and control patients were 60.4±8.5 and 62.5±7.7 years, respectively. Out of 50 CLL patients, 31 were male and 19 were female. The control group consisted of 32 male and 18 female. According to the Rai staging system, there were 32 (64.0%) patients with advanced Rai stage (≥ stage II) and 18 (36.0%) patients with limited Rai stage (< stage II).18 The demographic and laboratory specifications of the CLL patients and control subjects are presented in Table 1. Regarding the methylation frequency, results indicated that the promoter DNA methylation of miR-129-2 gene was more common among CLL patients than control subjects (38.0% vs. 0.0%; p < 0.001; χ2 = 23.457).
Table 1: The demographic and laboratory characteristics of study population.
|
Age, years, mean ± SD |
60.4 ± 8.5 |
62.5 ± 7.7 |
0.204 |
|
Sex (male/female) |
31/19 |
32/18 |
0.836 |
|
WBC, × 103/µL, mean ± SD |
74.0 ± 69.0 |
7.0 ± 1.0 |
< 0.001 |
|
Hb, g/dL, mean ± SD |
11.8 ± 2.87 |
13.8 ± 1.6 |
< 0.001 |
|
Plt, × 103/µL, mean ± SD |
137.0 ± 125.0 |
221.0 ± 569.0 |
< 0.001 |
|
Rai stage |
|
|
|
|
≥ 2 |
32 (64.0%) |
- |
- |
CLL: chronic lymphocytic leukemia; SD: standard deviation; WBC: white blood cell; Hb: hemoglobin; plt: platelet.
Table 2: Correlation between hypermethylation of miR-129-2 gene with clinical and laboratory features of chronic lymphocytic leukemia patients.
|
p-value |
0.190 |
0.708 |
0.122 |
0.020 |
0.086 |
0.019 |
WBC: white blood cell; Hb: hemoglobin; Plt: platelet.
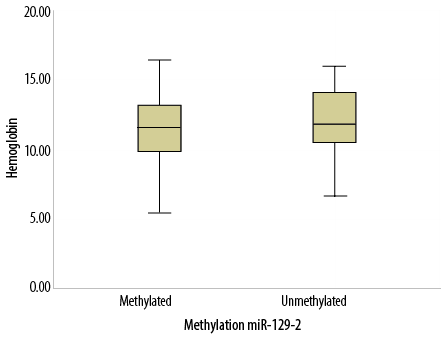
Figure 1: Box plot representing the correlation between hemoglobin levels and methylation of
miR-129-2 gene.
We evaluated the frequency of promoter DNA methylation of miR-129-2 gene between men and women to investigate possible gender-specific effects. The results indicated that promoter DNA methylation of the miR-129-2 gene was more common among men than women (14/31, 45.2% vs. 5/19, 26.3%). However, the difference was not statistically significant (p = 0.236; odds ratio = 2.30; 95% confidence interval: 0.63–7.37).
Moreover, the correlation of methylation status of the miR-129-2 gene with some clinical, laboratory, and immunological features of CLL patients revealed a significant but weak positive correlation between organomegaly and the methylated state of miR-129-2 gene (p = 0.019; r = 0.330). Organomegaly was significantly more common among CLL patients with a methylated rather than unmethylated miR-129-2 genes. Statistical analysis revealed a significant but weak negative correlation between hemoglobin levels and methylated state of miR-129-2 gene (p = 0.020;
r = -0.233) [Table 2]. Correlation analysis indicated that CLL patients with the methylated miR-129-2 gene had significantly lower hemoglobin levels than CLL patients with the unmethylated miR-129-2 gene [Figure 1]. However, we did not find any significant correlation between the methylation status of miR-129-2 gene and sex, age, WBC count, platelet count, and immunological markers CD2, CD3, CD5, CD19, CD20, CD23, and CD200 (p > 0.050).
Moreover, binary logistic regression analysis was done to investigate the independent association of each clinical and laboratory markers with the methylation status of miR-129-2 gene. Results indicated that organomegaly was the only marker with a significant association with hypermethylation of miR-129-2 gene (p = 0.046).
Discussion
We investigated the frequency of DNA methylation of miR-129-2 in a group of CLL patients. The main findings were as follows: (1) the DNA methylation frequency of miR-129-2 was significantly higher in the CLL group than the control group (p < 0.001), (2) CLL patients with methylated miR-129-2 displayed a higher frequency of organomegaly than those with unmethylated miR-129-2 (p = 0.019), (3) correlation analysis indicated a significant but weak correlation between organomegaly and hemoglobin levels with the methylated state of the miR-129-2 gene, and (4) binary logistic regression analysis between CLL patients with methylated and unmethylated miR-129-2 gene indicated organomegaly as the only associated significant factor (p = 0.046).
CLL presents as a cancer of elderly persons, and the role of environmental risk factors in its pathogenesis is not well characterized. Identification of genetic and epigenetic alterations may have pivotal roles in the diagnosis, prognosis, and treatment of CLL. Recently, special attention has been given to the role of micro RNA in the pathogenesis of CLL. Our study reported a higher frequency of miR-129-2 methylation in patients with CLL in accordance with previously published studies.19 However, investigation of the frequency of miR-129-2 methylation in myeloid malignancies did not display such an association.15 So it seems that epigenetic silencing of miR-129-2 has a tumor-specific role. The conflicting finding regarding the methylation of miR-129-2 in lymphoid (CLL) and myeloid cancers may be explained by the presence of gene-gene, gene-environmental, and epigenetic interactions.20,21
Aberrant hypermethylation of miR-129-2 has been reported in several cancers, including stomach, lung, and glioma.13,14,22 In a 2019 study, the methylation status of miR-129-2 was evaluated using the MSP method in 50 gastric cancer patients and the results indicated aberrant methylation in 84% of patients.22 They concluded that miR-129-2 methylation might play a pivotal role in the progression of gastric cancer.22
Another study investigated the methylation status of miR-129-2 in five human lung cancer cell lines and indicated that miR-129-2 was absolutely methylated in A549, SPCA-1, SK-MES-1, and PC-9 cells and unmethylated in 95-D cells suggesting hypermethylation of miR-129-2 in the majority of lung carcinoma.13
These studies represent the central role of miR-129-2 methylation in the development of numerous cancers. CLL is a disease with a high prevalence and intensity in men than women. We explored if the higher intensity of CLL disease in men than women could be attributed to a different rate of DNA methylation between the two sexes.23 DNA methylation of miR-129-2 was more common in males than females (45.2% vs. 26.3%); however, the difference was not statistically significant. As our study sample size was small, the replication of the study with a larger sample size may elucidate the exact effect of this epigenetic change on the severity of CLL disease in men. It has been suggested that differences in DNA methylation frequency may contribute to the sex-related difference in CLL risk.24
Based on the Rai classification system, splenomegaly, hepatomegaly, and lymphadenopathy contribute to the staging of CLL disease.25 Organomegaly is associated with the intermediate severity of CLL disease. Our results indicated a higher prevalence of methylated miR-129-2 in CLL patients with organomegaly relative to CLL patients without organomegaly (p = 0.033; χ2 = 5.433), which indicate a possible role for the methylated miR-129-2 gene in determining the severity of CLL disease. So, the methylated miR-129-2 gene may act as an epigenetic predictor of disease severity. However, this preliminary result needs to be verified in future studies with a preferably prospective study design. Our study bears some limitations, including: (1) the sample size was relatively small, (2) the gene expression of miR-129-2 was not determined, and (3) the study design was retrospective.
Conclusion
DNA methylation of miR-129-2 may be involved in the development of CLL. Moreover, the methylated miR-129-2 gene may act as an epigenetic predictor of CLL disease. A prospectively designed study with a large sample size may be necessary for better understanding of the role of miR-129-2 in pathogenesis and prognosis of CLL disease.
Disclosure
The authors declared no conflict of interest. No funding was received for this study. The research is the result of a master’s thesis in laboratory hematology and blood bank approved by the Research Center, High Institute for Research and Education in Transfusion Medicine.
references
- 1. Hallek M, Shanafelt TD, Eichhorst B. Chronic lymphocytic leukaemia. Lancet 2018 Apr;391(10129):1524-1537.
- 2. Karan-Djurasevic T, Pavlovic S. Somatic hypermutational status and gene repertoire of immunoglobulin rearrangements in chronic lymphocytic leukemia. lymphocyte updates: cancer. Autoimmunity and Infection 2017 Jul;49.
- 3. Agathangelidis A, Ljungström V, Scarfò L, Fazi C, Gounari M, Pandzic T, et al. Highly similar genomic landscapes in monoclonal B-cell lymphocytosis and ultra-stable chronic lymphocytic leukemia with low frequency of driver mutations. Haematologica 2018 May;103(5):865-873.
- 4. Hillmen P. Using the biology of chronic lymphocytic leukemia to choose treatment. Hematology Am Soc Hematol Educ Program 2011;2011(1):104-109.
- 5. Guièze R, Wu CJ. Genomic and epigenomic heterogeneity in chronic lymphocytic leukemia. Blood 2015 Jul;126(4):445-453.
- 6. Parikh SA, Shanafelt TD. Prognostic factors and risk stratification in chronic lymphocytic leukemia. Semin Oncol 2016 Apr;43(2):233-240.
- 7. Bagacean C, Tempescul A, Le Dantec C, Bordron A, Mohr A, Saad H, et al. Alterations in DNA methylation/demethylation intermediates predict clinical outcome in chronic lymphocytic leukemia. Oncotarget 2017 Aug;8(39):65699-65716.
- 8. Mirzaei H, Fathullahzadeh S, Khanmohammadi R, Darijani M, Momeni F, Masoudifar A, et al. State of the art in microRNA as diagnostic and therapeutic biomarkers in chronic lymphocytic leukemia. J Cell Physiol 2018 Feb;233(2):888-900.
- 9. Iorio MV, Croce CM. MicroRNAs in cancer: small molecules with a huge impact. J Clin Oncol 2009 Dec;27(34):5848-5856.
- 10. Pei L, Choi JH, Liu J, Lee EJ, McCarthy B, Wilson JM, et al. Genome-wide DNA methylation analysis reveals novel epigenetic changes in chronic lymphocytic leukemia. Epigenetics 2012 Jun;7(6):567-578.
- 11. Chen X, Zhang L, Zhang T, Hao M, Zhang X, Zhang J, et al. Methylation-mediated repression of microRNA 129-2 enhances oncogenic SOX4 expression in HCC. Liver Int 2013 Mar;33(3):476-486.
- 12. Shen R, Pan S, Qi S, Lin X, Cheng S. Epigenetic repression of microRNA-129-2 leads to overexpression of SOX4 in gastric cancer. Biochem Biophys Res Commun 2010 Apr;394(4):1047-1052.
- 13. Xiao Y, Li X, Wang H, Wen R, He J, Tang J. Epigenetic regulation of miR-129-2 and its effects on the proliferation and invasion in lung cancer cells. J Cell Mol Med 2015 Sep;19(9):2172-2180.
- 14. Yang Y, Huang JQ, Zhang X, Shen LF. MiR-129-2 functions as a tumor suppressor in glioma cells by targeting HMGB1 and is down-regulated by DNA methylation. Mol Cell Biochem 2015 Jun;404(1-2):229-239.
- 15. Wong KY, Yim RL, Kwong YL, Leung CY, Hui PK, Cheung F, et al. Epigenetic inactivation of the MIR129-2 in hematological malignancies. J Hematol Oncol 2013 Feb;6(1):16.
- 16. Suguna S, Nandal D, Kamble S, Bharatha A, Kunkulol R. Genomic DNA isolation from human whole blood samples by non enzymatic salting out method. Int J Pharm Pharm Sci 2014 May;6(6):198-199.
- 17. Liang Y, Yang X, Ma L, Cai X, Wang L, Yang C, et al. Homocysteine-mediated cholesterol efflux via ABCA1 and ACAT1 DNA methylation in THP-1 monocyte-derived foam cells. Acta Biochim Biophys Sin (Shanghai) 2013 Mar;45(3):220-228.
- 18. Hallek M. Chronic lymphocytic leukemia: 2015 update on diagnosis, risk stratification, and treatment. Am J Hematol 2015 May;90(5):446-460.
- 19. Kunej T, Godnic I, Ferdin J, Horvat S, Dovc P, Calin GA. Epigenetic regulation of microRNAs in cancer: an integrated review of literature. Mutat Res 2011 Dec;717
(1-2):77-84.
- 20. Coyle KM, Boudreau JE, Marcato P. Genetic mutations and epigenetic modifications: driving cancer and informing precision medicine. Biomed Res Int 2017;2017:9620870.
- 21. Talukdar FR, Ghosh SK, Laskar RS, Mondal R. Epigenetic, genetic and environmental interactions in esophageal squamous cell carcinoma from northeast India. PLoS One 2013 Apr;8(4):e60996.
- 22. Alizadeh N, Asadi M, Shanehbandi D, Zafari V, Shomali N, Asvadi T, et al. Evaluation of the methylation of MIR129-2 gene in gastric cancer. J Gastrointest Cancer 2019 May;51(6):1-4.
- 23. Catovsky D, Wade R, Else M. The clinical significance of patients’ sex in chronic lymphocytic leukemia. Haematologica 2014 Jun;99(6):1088-1094.
- 24. Lin S, Liu Y, Goldin LR, Lyu C, Kong X, Zhang Y, et al. Sex-related DNA methylation differences in B cell chronic lymphocytic leukemia. Biol Sex Differ 2019 Jan;10(1):2.
- 25. Rai KR, Stilgenbauer S. Staging and prognosis of chronic lymphocytic leukemia. 2019 [cited 2019 Dec 3]. Available from: https://www.uptodate.com/contents/staging-and-prognosis-of-chronic-lymphocytic-leukemia.