| |
Introduction
Lactose, the principle sugar in milk is a disaccharide, hydrolyzed by intestinal lactase (EC3.2.1.23/26) into glucose and galactose, which are absorbed directly by diffusion in the intestine. The decline of lactase expression in intestinal microvilli after weaning is a normal phenomenon in mammals known as lactase non-persistence (LNP).1 In adults, the activity of lactase is low and lactose cannot be hydrolyzed, resulting in malabsorption. Lactose malabsorption (LM) is defined as a failure of the mechanisms of absorption of lactose that can be shown by laboratory tests. The term lactose intolerance (LI) is used for a clinical entity, describing symptomatic lactose malabsorption such as bloating, flatulence, abdominal pain and diarrhoea.2,3
There are various methods used to assess the absorption of lactose in the digestive system such as the lactose tolerance test, lactose hydrogen breath test (LHBT), stool acidity tests and intestinal biopsy. Among these methods, LHBT is the test of choice because it is non-invasive and has good sensitivity and specificity. In adults with LI, bacterial microflora ferments lactose and releases hydrogen, methane and different amino acids.4 The H2 diffuses into the blood circulation and is exhaled by the lung.5 LHBT is based on measuring the level of H2 production after ingesting lactose. It serves as one of the reliable methods for diagnosing LM and LI.6-8
In humans, the lactase gene (LPH or LCT) is located on chromosome 2q21. It comprises 17 exons and covers approximately 49 kb. Investigation of the lactase activity showed that several single nucleotide polymorphisms [SNPs] (C/G-13907, C/T-13910, T/C-13913, G/A-13914, T/G-13915, G/C-14010) within the MCM6 gene act upstream of the lactase gene, allowing lactase gene expression and maintain lactase activity in adults. These SNPs have strong association with lactase persistence in different populations. Individuals with one of the above mentioned SNPs, usually maintain high to normal activity of lactase throughout their adult lives, a condition known as lactase persistence (LP).9 In Arabs of Oman, Yemen and Saudi Arabia, the G-13915 is the most common LP-associated allele in contrast to the T-13910 in Caucasians.10-16
The aim of the present study was to investigate the phenotype-genotype correlation of lactase persistence in Omani adults. The LHBT was used for phenotyping; while direct DNA sequencing was used for genotyping.
Methods
This study was conducted at Sultan Qaboos University, Muscat, Oman from January 2008- February 2009. The protocol was reviewed and approved by the Medical Research and Ethics Committee at the College of Medicine. All subjects signed a written informed consent for participation into the study. Fifty-two university students and staff (31 males, 21 females) from different regions of the Sultanate of Oman volunteered for the study with a mean age ± SD of 25 ± 5.4 years, ranging from 19-42 years.
For the Lactose Hydrogen Breath Test (LHBT), participants were instructed to fast overnight for at least 10 hours before the LHBT. The test was performed after oral ingestion of 50 g lactose diluted in 250 ml of water. Breath H2 excretion was measured in parts per million (ppm) using a Micro H2 Analyzer (Micro Medical Limited, Chatman, UK) which detects hydrogen concentrations in the range of 0-500 ppm. The measurements were taken at baseline and every 30 minutes after the ingestion of lactose over a period of 180 minutes. The LHBT is considered positive (lactose malabsorption) when H2 excretion is higher than 20 ppm above the baseline or nadir level.17 In addition, the occurrence of gastrointestinal (GI) symptoms such as abdominal pain, bloating, diarrhoea, flatulence and gases during or after the test was documented.
For Genotyping, DNA was isolated from blood samples of the volunteers by phenol-chloroform extraction. A DNA fragment (383bp) in the MCM6 gene spanning the C/T-13910 SNP in Intron 13 was amplified according to Ingram et al.14 with modified primers: (LAC13F: 5`-GAATGCTCATACGACCATGGAA-3` and LAC13R: 5`-CTGCTTTGGTTGAAGCGAAG-3`), (Metabion, Martinsried, Germany). Polymerase chain reaction (PCR) amplification was carried out in a 20 μL volume with genomic DNA (100 ng), primers (20 ng each), dNTPs (200 μM) and 0.5 U of Taq polymerase in a standard buffer (Applied Biosystems, NY, USA). The PCR cycle conditions were as follows: an initial round of denaturation at 94ºC, then 35 cycles at 94ºC for 30 seconds, 58ºC for 30 seconds, 72ºC for 75 seconds, and a final extension of 72ºC for 10 minutes.
The amplicon (PCR product) was purified by ExoSAP (Applied Biosystems, Ohio, USA) and the sequencing reaction was performed using the Big Dye terminator (version 1.1). The products were then loaded into ABIPrizm 3100 genetic analyzer (Applied Biosystems, CA, USA).
The chromatograms of genetic analysis were read using Lasergene 7.0 software. Data were statistically analyzed using the SPSS software, version 19.0. The agreement between LHBT and molecular analysis was assessed. Specificity, sensitivity, accuracy and predictive value of genotyping versus phenotyping were estimated; while, the kappa coefficient (95% CI), was determined with the level of significance set at 5%.
Results
Of the 52 participants, 42 (81%) had positive LHBT and were considered LM. Of those, 32 (78%) had gastro-intestinal (GI) symptoms (Fig. 1). Negative tests were observed in 8 (15%) individuals, three of whom had GI symptoms, while two (4%) individuals were defined as non-hydrogen producers but also had symptoms.
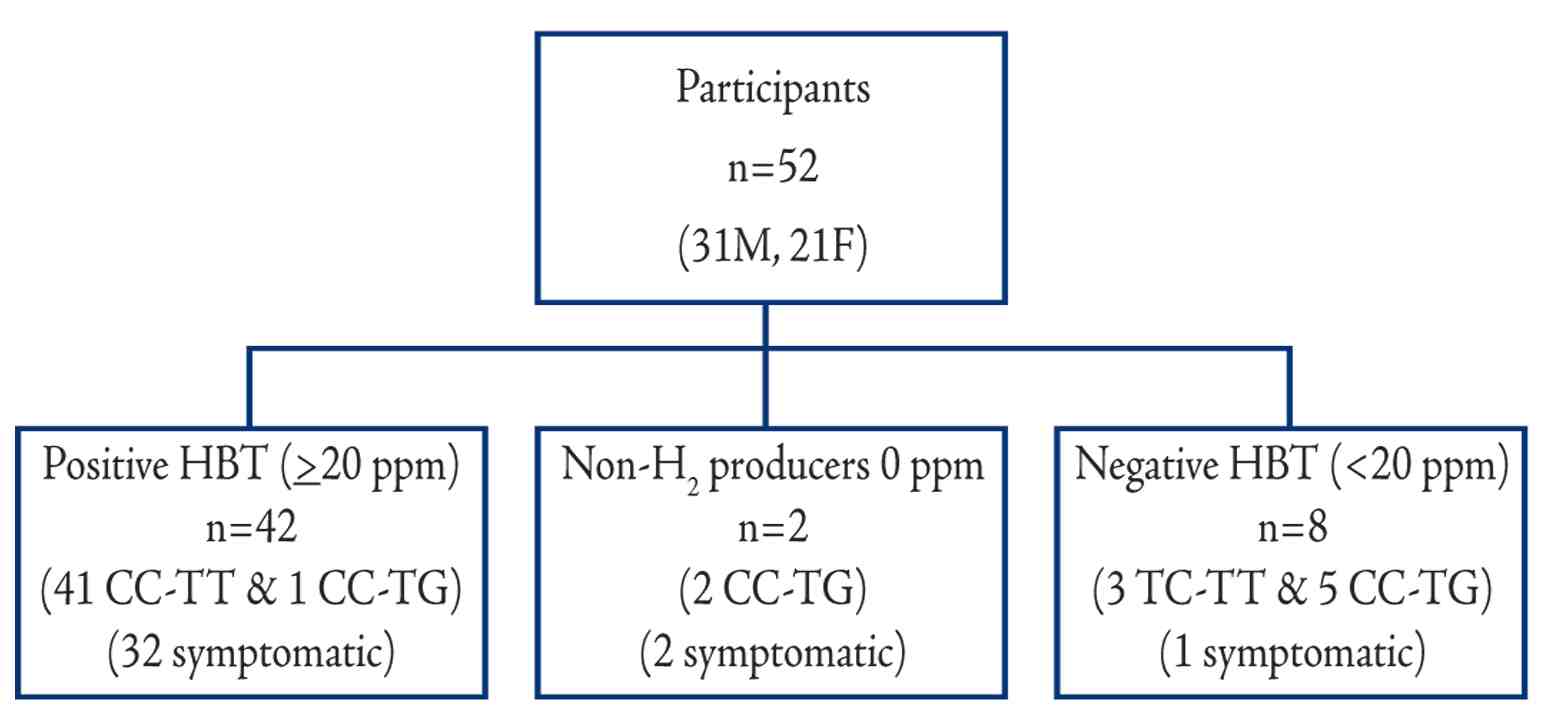
Figure 1: Schematic flow chart of Omani participants showing results of LHBT, genotypes, and GI symptoms.
Only two lactase associated SNPs, the T-13910 and the G-13915 were detected among Omani people. All the 42 (100%) individuals genotyped with the CC-13910 and TT-13915 had positive LHBT and 30 (73%) of them developed GI symptoms. All the three individuals with T/C-13910 and seven out of the eight genotyped as T/G-13915, had negative LHBT (Table 1). Three participants (one carried T-13910 and two G-13915) out of the ten with LP-alleles had GI symptoms. There was a highly significant difference (p<0.001) in the average hydrogen levels between the lactose malabsorbers-group (genotyped as wild type) and the absorbers with mutated genotype (either T-13910 or G-13915). (Fig. 2)
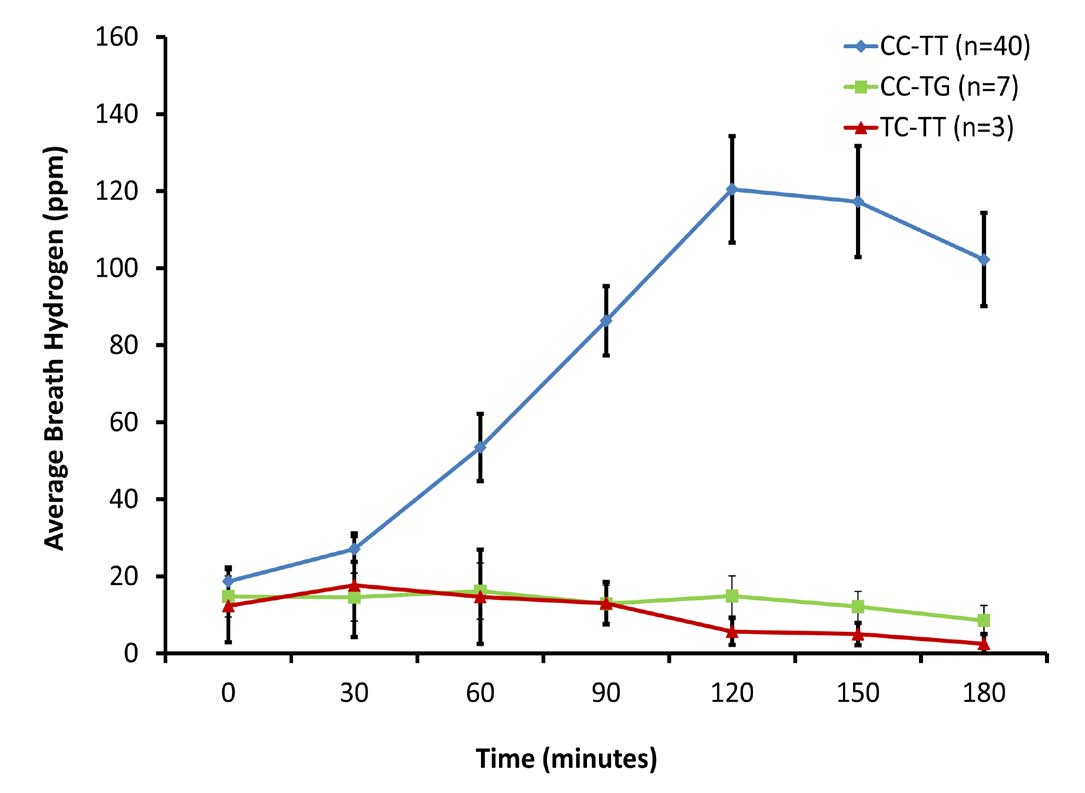
Figure 2: Lactose Hydrogen Breath Test among the three genotyped groups.
Table 1: Association of genotypes (CT-13910 & TG-13915) with LHBT results among Omani participants.
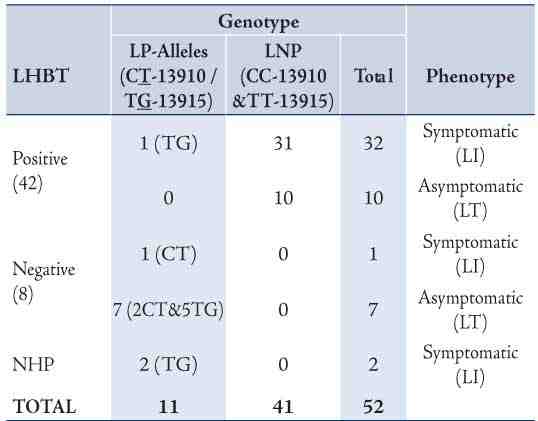
The results revealed significant agreement between the C/T-13910 (Indo-Asian and Caucasian allele) and the T/G-13915 (Arab-specific allele) genotype with LHBT (Kappa index 0.9). Individuals with either T-13910 or G-13915 allele (lactase persistence) had significantly lower breath H2 peak (Average: 8 and 10 ppm, respectively) compared to the C/C-13910 and T/T-13915 (Average: 121 ppm). All the 9 lactose absorbers (H2 <20 ppm), had either G-13915 or T-13910. In contrast, among those 42 participants diagnosed as lactose malabsorbers, (H2 ≥20 ppm), 41 were homozygous C/C-13910 and T/T-13915. However, a 21 year-old female participant who carried the T/G-13915 variant had a positive LHBT (37 ppm) and did not develop symptoms during the test. We did not investigate whether she had any secondary GI disorder or disease that might have explained this anomaly.
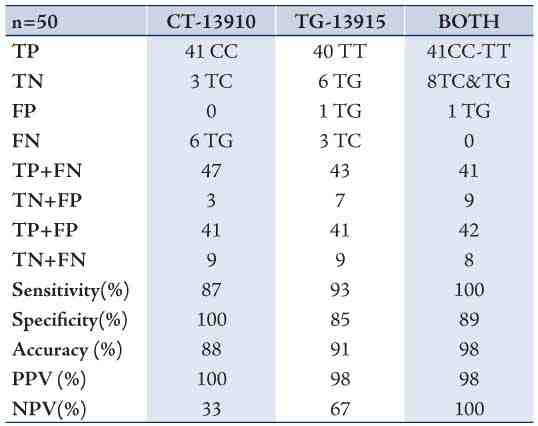
Using the LHBT as a standard method, genotyping exhibited 100% sensitivity, 89% specificity, 98% accuracy, 98% positive predictive value and 100% negative predictive value in the tested adult population when both SNPs were tested. (Table 2)
Table 2: LHBT in comparison with CT-13910 and TG-13915 genotypes (singly and both).
Discussion
From the study findings and from other studies, it is apparent to us and other investigators that GI symptoms are not always a complete indicator of lactose intolerance.18,19 Only 32/42 (76%) of malabsorbers (positive LHBT) reported symptoms, while 1/8 (13%) of lactose absorbers (negative LHBT) also reported symptoms (Fig. 1). In other words, not all lactose malabsorbers are lactose intolerant. This cannot be fully explained. In many cases, adaptation associated with different types of intestinal microflora or other secondary causes may explain the discordance.
Identification of LP polymorphisms is complementary to LHBT and adds value to screening, diagnosis and management of LI. In this study, several mutations and/or variants (C/G-13907, C/T-13910, T/C-13913, T/G-13915, G/C-14010, G/A-22018, and T/C-3712) were evaluated and tested against lactase persistence. Of these SNPs, only two (C/T-13910 and T/G-13915) seemed to be associated with lactase persistence among Omani individuals.
The correlation and different patterns of LHBT have been investigated extensively for the C/T-13910 polymorphism.20-24 However, there is only one study in the literature where two subjects carrying the T/G-13915 had hydrogen levels <20 ppm which is in concordance with our results.24 Specificity, Sensitivity, accuracy, positive and negative predictive values improved when both SNPs (C/T-13910 and T/G-13915) were genotyped reaching 100, 89, 98, 98 and 100%; respectively in this study. When one SNP is tested in mixed populations, misdiagnosis (false negatives) seems to increase. Secondary causes of hypolactasia should also be excluded, if the genotype is not in concordance with LHBT, in order to complete the diagnosis of lactose intolerance. It is important to stress here that neither C/T-13910 nor G/T-13915 alone can explain the pattern of LI among Omanis.
Conclusion
The highest degree of sensitivity, specificity, accuracy and predictive values for diagnosing LP can only be achieved when all SNPs associated with it are investigated, especially in ethnically mixed populations (i.e., at least both C/T-13910 and G/T-13915 must be tested in an Omani population). Genotyping by itself can only be used as a complementary test, with LHBT as a primary test for LI, especially in adults. However, genotyping can be used as a screening and even as a predictive tool among children.
Acknowledgements
The authors reported no conflict of interest. This work was funded by Sultan Qaboos University under grant No. IG/MED/BIOC/10/02.
References
1. McBean LD, Miller GD. Allaying fears and fallacies about lactose intolerance. J Am Diet Assoc 1998 Jun;98(6):671-676.
2. Montalto M, Curigliano V, Santoro L, Vastola M, Cammarota G, Manna R, et al. Management and treatment of lactose malabsorption. World J Gastroenterol 2006 Jan;12(2):187-191.
3. Sahi T, Isokoski M, Jussila J, Launiala K, Pyörälä K. Recessive inheritance of adult-type lactose malabsorption. Lancet 1973 Oct;2(7833):823-826.
4. Lomer MC, Parkes GC, Sanderson JD. Review article: lactose intolerance in clinical practice–myths and realities. Aliment Pharmacol Ther 2008 Jan;27(2):93-103.
5. Levitt MD. Production and excretion of hydrogen gas in man. N Engl J Med 1969 Jul;281(3):122-127.
6. Usai-Satta P, Scarpa M, Oppia F, Cabras F. Lactose malabsorption and intolerance: What should be the best clinical management? World J Gastrointest Pharmacol Ther 2012 Jun;3(3):29-33.
7. Corazza GR, Sorge M, Strocchi A, Lattanzi MC, Benati G, Gasbarrini G. Methodology of the H2 breath test. II. Importance of the test duration in the diagnosis of carbohydrate malabsorption. Ital J Gastroenterol 1990 Oct;22(5):303-305.
8. Romagnuolo J, Schiller D, Bailey RJ. Using breath tests wisely in a gastroenterology practice: an evidence-based review of indications and pitfalls in interpretation. Am J Gastroenterol 2002 May;97(5):1113-1126.
9. Troelsen JT. Adult-type hypolactasia and regulation of lactase expression. Biochim Biophys Acta. 2005;1723:19-32
10. Al-Abri AR, Al-Rawas O, Al-Yahyaee S, Al-Habori M, Al-Zubairi AS, Bayoumi RA. Distribution of the lactase persistence-associated variant alleles -13910* T and -13915* G among the people of Oman and Yemen. Hum Biol 2012;84(3):271-286.
11. Enattah NS, Jensen TG, Nielsen M, Lewinski R, Kuokkanen M, Rasinpera H, et al. Independent introduction of two lactase-persistence alleles into human populations reflects different history of adaptation to milk culture. Am J Hum Genet 2008 Jan;82(1):57-72.
12. Enattah NS, Sahi T, Savilahti E, Terwilliger JD, Peltonen L, Järvelä I. Identification of a variant associated with adult-type hypolactasia. Nat Genet 2002 Feb;30(2):233-237.
13. Imtiaz F, Savilahti E, Sarnesto A, Trabzuni D, Al-Kahtani K, Kagevi I, et al. The T/G 13915 variant upstream of the lactase gene (LCT) is the founder allele of lactase persistence in an urban Saudi population. J Med Genet 2007 Oct;44(10):e89.
14. Ingram CJ, Elamin MF, Mulcare CA, Weale ME, Tarekegn A, Raga TO, et al. A novel polymorphism associated with lactose tolerance in Africa: multiple causes for lactase persistence? Hum Genet 2007 Feb;120(6):779-788.
15. Mulcare CA, Weale ME, Jones AL, Connell B, Zeitlyn D, Tarekegn A, et al. The T allele of a single-nucleotide polymorphism 13.9 kb upstream of the lactase gene (LCT) (C-13.9kbT) does not predict or cause the lactase-persistence phenotype in Africans. Am J Hum Genet 2004 Jun;74(6):1102-1110.
16. Tishkoff SA, Reed FA, Ranciaro A, Voight BF, Babbitt CC, Silverman JS, et al. Convergent adaptation of human lactase persistence in Africa and Europe. Nat Genet 2007 Jan;39(1):31-40.
17. Joseph F Jr, Rosenberg AJ. Breath hydrogen testing: diseased versus normal patients. J Pediatr Gastroenterol Nutr 1988 Sep-Oct;7(5):787-788.
18. Beyerlein L, Pohl D, Delco F, Stutz B, Fried M, Tutuian R. Correlation between symptoms developed after the oral ingestion of 50 g lactose and results of hydrogen breath testing for lactose intolerance. Aliment Pharmacol Ther 2008 Apr;27(8):659-665.
19. Gugatschka M, Dobnig H, Fahrleitner-Pammer A, Pietschmann P, Kudlacek S, Strele A, et al. Molecularly-defined lactose malabsorption, milk consumption and anthropometric differences in adult males. QJM 2005 Dec;98(12):857-863.
20. Büning C, Genschel J, Jurga J, Fiedler T, Voderholzer W, Fiedler EM, et al. Introducing genetic testing for adult-type hypolactasia. Digestion 2005;71(4):245-250.
21. Högenauer C, Hammer HF, Mellitzer K, Renner W, Krejs GJ, Toplak H. Evaluation of a new DNA test compared with the lactose hydrogen breath test for the diagnosis of lactase non-persistence. Eur J Gastroenterol Hepatol 2005 Mar;17(3):371-376.
22. Pohl D, Savarino E, Hersberger M, Behlis Z, Stutz B, Goetze O, et al. Excellent agreement between genetic and hydrogen breath tests for lactase deficiency and the role of extended symptom assessment. Br J Nutr 2010 Sep;104(6):900-907.
23. Bulhões AC, Goldani HA, Oliveira FS, Matte US, Mazzuca RB, Silveira TR. Correlation between lactose absorption and the C/T-13910 and G/A-22018 mutations of the lactase-phlorizin hydrolase (LCT) gene in adult-type hypolactasia. Braz J Med Biol Res 2007 Nov;40(11):1441-1446.
24. Haberkorn BC, Ermens AA, Koeken A, Cobbaert CM, van Guldener C. Improving diagnosis of adult-type hypolactasia in patients with abdominal complaints. Clin Chem Lab Med 2012 Jan;50(1):119-123.
|
|